New tube maintains EVs and cfRNA
Topics Featured
Streck R&D Senior Scientific Manager Nick George, Ph.D., recently presented a 15-minute talk titled “Introducing RNA Complete BCT® (RUO), a novel blood collection tube that maintains extracellular vesicles and cell-free RNA for applications like next generation sequencing” at the SelectScience Virtual Summit on Cancer and Immunology Research.
RNA Complete BCT is a direct draw blood collection tube that maintains draw time concentrations of both cell-free RNA (cfRNA) and extracellular vesicles (EV). The tube maintains both of these critical populations for up to 7 days at room temperature, maximizing plasma yield and minimizing hemolysis during the storage time. This tube is potentially useful to those interested in assaying EVs and cell-free RNA.
Liquid biopsy research assays targeting EVs and cfRNA provide a comprehensive and dynamic view of the disease state. Dr. George describes the Streck RNA Complete BCT’s incorporation into a workflow allowing for delayed sample processing without effect on sample integrity or downstream analyte analysis in research applications. Data includes information generated from next generation sequencing, ddPCR and EV counting by Malvern NanoSight.
Watch this presentation to learn more about:
- The need in liquid biopsy research for sample stabilization of extracellular vesicles and cell-free RNA
- The performance of RNA Complete BCT in maintaining draw time levels of EVs and cell-free RNA
- The use of RNA Complete BCT in RNA next generation sequencing workflows
Video transcription
To those of you attending this presentation, today I will be describing a blood collection tube, RNA Complete, recently launched by our group at Streck. The RNA Complete blood collection tube is for research use only and is not intended for diagnostic procedures.
I’ll begin by giving a very brief background on Streck and our widely used cell-free DNA blood collection tube. From here, I will transition to an area of unmet need in liquid biopsy, specifically sample stabilization of extracellular vesicles, or EVs, and EV associated cell-free RNA. The performance of RNA Complete, Streck’s answer to this area of unmet need, will then be described, specifically maintaining draw time levels of EVs and cell-free RNA. Finally, I’ll describe our future directions with this blood collection tube.
Streck’s background in preanalytics
Streck was founded in 1971, and we celebrate our 50th anniversary this coming year. Our core technology is cell stabilization, and this knowledge is exploited in the manufacture of hematology, flow cytometry, and other biofluid controls. Most familiar to those in this audience is our cell-free DNA blood collection tube, first introduced in 2010. The Streck Cell-Free DNA BCT® stabilizes nucleated blood cells, thus preventing a release of genomic DNA with blood storage time. This prevents non-specific increases in the cell-free DNA fraction, which is critical to assay by those in the liquid biopsy research field. Intended use of Cell-Free DNA BCT is both cell-free DNA and circulating tumor cell research.
The need for extracellular vesicle and cfRNA sample stabilization
Body fluids samples, such as blood, contain many other analytes in addition to cell-free DNA that are useful to liquid biopsy assays. These include extracellular vesicles and EV associated cell-free RNA. Other analytes include non-EV associated cell-free RNA; for example, certain microRNAs and protein or protein complexes useful in proteomic analysis.
What are extracellular vesicles?
A little bit of background on extracellular vesicles. EVs are a heterogenous population of particles found in nearly all body fluids, and characterized by both size and membrane protein marker expression. EVs are broadly grouped as larger microvesicles, which bleb passively from the host plasma membrane and the smaller exosomes, which are actively secreted by the host cell. Why do we care about EVs? Because first, these vesicles are released by all cells. Thus, all cell types in a collected blood sample can and do release EVs. This becomes problematic for assays aimed at characterizing EV populations or EV contents, such as messenger RNA or microRNA, as cellular breakdown may lead to non-specific increases in these analytes.
Current preanalytical product limitations
What are the currently available options for EV and cell-free RNA stabilization? Well, there are a multitude of BCTs intended for use in cell-free DNA, but these specifically target white blood cells, thus preventing a release in genomic DNA. There is little concern with other cell types, such as red cells and platelets, cells that release EVs and contain abundant RNA.
What happens to blood samples stored in common EDTA blood collection tubes?
We began our studies by characterizing what happens to blood samples stored in common BCTs, such as EDTA blood collection tubes. During ambient temperature storage, these samples displayed time dependent increases in hemolysis, as demonstrated in the left and center panels (Figure 1). Quantitative increases in hemolysis are most apparent between 3 and 7 days of blood storage in these tubes. A parallel increase in EVs is also observed in these samples, as demonstrated on the right panel (Figure 1).
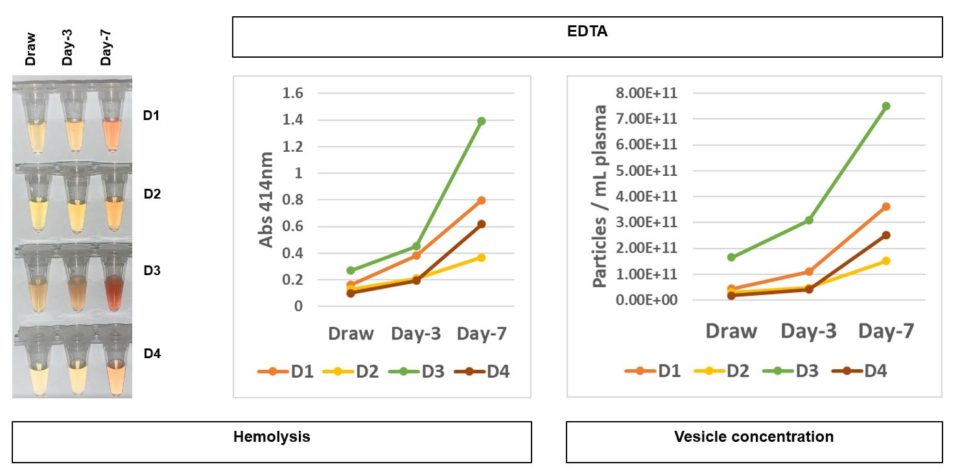
Figure 1
What happens to blood samples stored in cfDNA blood collection tubes?
Next we compared observations with samples drawn into EDTA BCTs with equivalent donor samples drawn into the Streck Cell-Free DNA BCT. Both tube types demonstrated robust storage time dependent increases in EVs, as shown in the bottom panels (Figure 2). Not similar between the two tube types was cell-free DNA concentration, as determined by droplet digital PCR analysis of beta-actin copy number. Here, EDTA samples demonstrated a strong increase in cell-free DNA, as shown in the top panel (Figure 2). In contrast, the Cell-Free DNA BCT maintained draw time concentrations of cell-free DNA. Thus, the Cell-Free DNA BCT performed its intended function, but should not be used for studies investigating EVs.
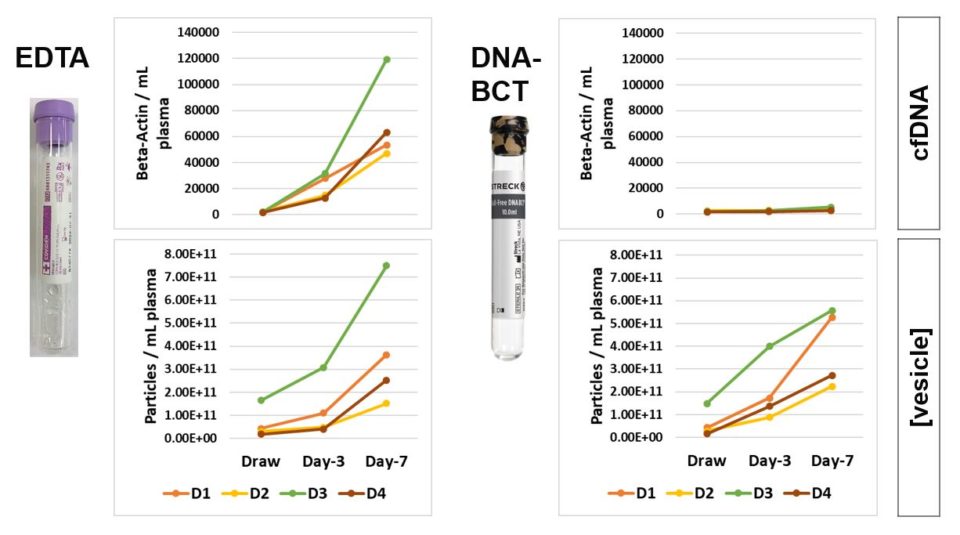
Figure 2
What is the effect on cell-free RNA concentration?
What about cell-free RNA? As anticipated based on increases in EV concentration, neither blood collection tube maintained draw time concentrations of cell-free RNA, as determined by bioanalyzer analysis for two different healthy donors. Draw time levels of cell-free RNA are denoted by the overlapping green and dark blue lines for EDTA and Cell-Free DNA BCT samples respectively (Figure 3).
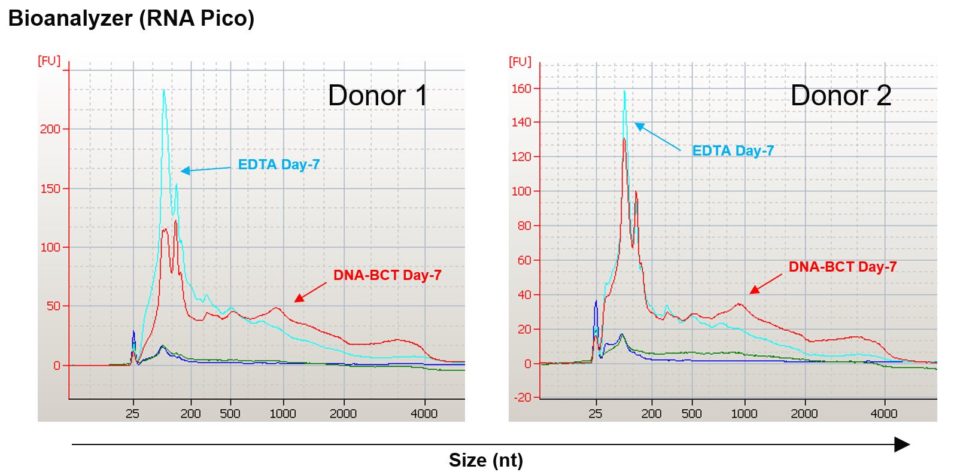
Figure 3
Why is this all important?
Why is all this important? Because many new age assays are leveraging a combined analyte approach. It was our goal to develop a blood collection tube that fills the current void for extracellular vesicle and cell-free RNA analysis. With that said, we introduce the Streck RNA Complete BCT. The Streck RNA Complete BCT is a direct draw blood collection tube that maintains the draw time concentration of cell-free RNA and extracellular vesicles, such as exosomes. Preservative stabilizes cell-free RNA and exosome counts for up to 7 days at room temperature, maximizing plasma yields and minimizing hemolysis during storage.
“It was our goal to develop a blood collection tube that fills the current void for extracellular vesicle and cell-free RNA analysis.”
In creating the RNA Complete BCT, we went about development according to the following strategy. The overall intent was to stabilize all cells in the collected blood sample, including white cells, mature and immature red cells, and platelets. In doing so, we would limit hemolysis, maintain draw time plasma volume, and prevent release of extracellular vesicles with ambient condition storage time. Because EVs are the protectant shield for the majority of plasma cell-free RNA, maintaining draw time concentrations of EVs would in turn result in stabilization of draw time cell-free RNA concentration.
Outlined here is our general workflow for testing the functionality of RNA Complete BCT. Healthy donors are drawn onsite, and plasma is either isolated immediately, day 0, or after ambient temperature storage; for example, on day 7. Plasma isolation utilizes a general double spin protocol. From here, plasma is stored frozen at -80°, or immediately utilized in assays. For extracellular vesicle analysis, we utilize the Malvern NS300 instrument. For cell-free RNA analysis, we first purified the plasma nucleic acid combined with the DNase1 digestion step. From here, cell-free RNA is reverse transcribed and then used in transcript specific analysis via droplet digital PCR. Alternatively, total cell-free RNA is analyzed via bioanalyzer or fluorometric means.
RNA Complete BCT performance
How does RNA Complete perform? First we looked at general whole blood parameters. As demonstrated in the left panels, visual hemolysis is minimal and appears similar to draw time levels (Figure 4). EDTA BCTs, containing the purple stoppers, demonstrate robust hemolysis after 7 days of ambient temperature storage. Hemolysis is quantitated by hemoglobin absorbents in the upper right panels. Finally, recoverable plasma volume was determined for both EDTA and RNA Complete BCTs. While EDTA blood demonstrated a roughly 45% decrease in recoverable plasma volume, the RNA Complete BCT maintained draw time plasma volume equating to roughly 1.5 milliliters more of plasma after 7 days ambient temperature storage. Plasma volume is related to changes in red blood cell mean corpuscular volume. This was also maintained to draw time levels in RNA Complete BCT, as shown in the bottom right panel (Figure 4).
RNA Complete BCT formulation limits hemolysis and maintains draw time plasma volume.
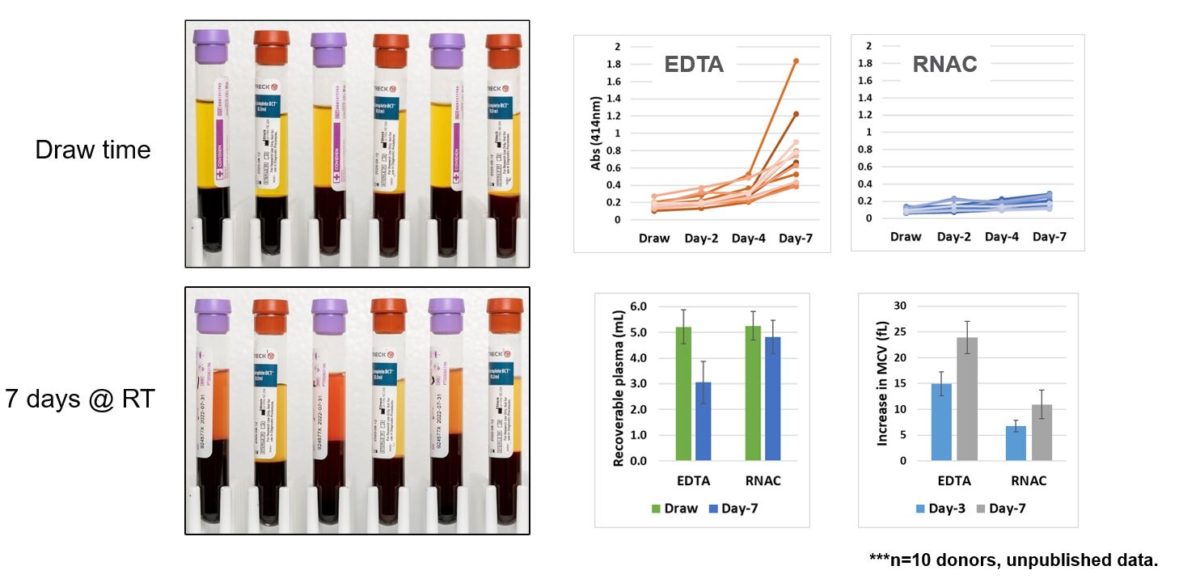
Figure 4
Next, we looked at extracellular vesicle concentration with storage time. Whereas EV concentration goes up in the control EDTA BCTs, EV concentration was maintained to near draw time levels in the RNA Complete BCT. Importantly, draw time concentrations of EVs are the same between EDTA and RNA Complete samples, as shown in the far right panel (Figure 5).
RNA Complete BCT formulation limits increases in EVs during ambient temperature storage.
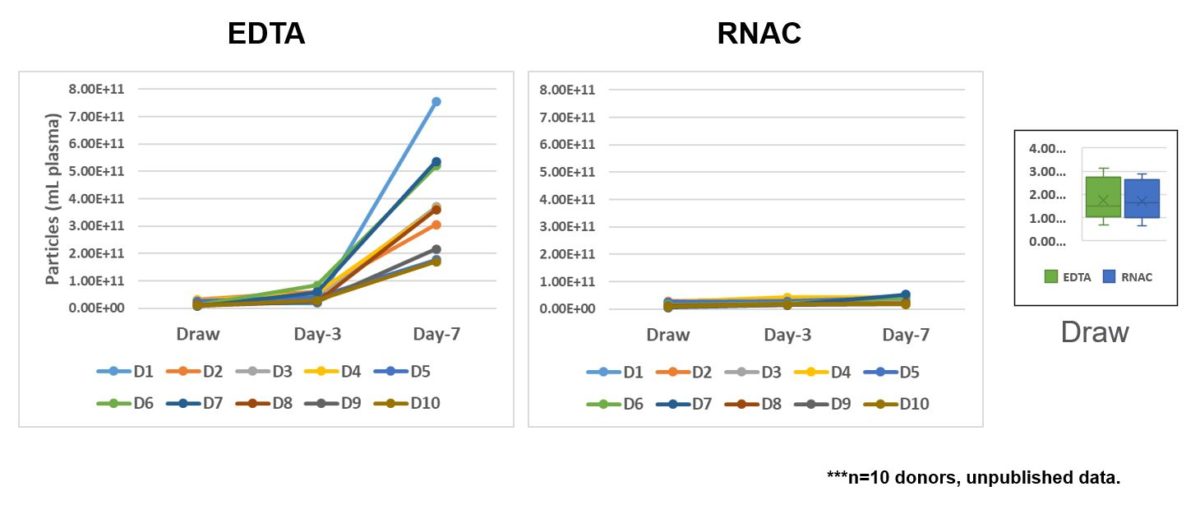
Figure 5
With knowledge that EV concentration is maintained over the course of 7 days, we next determined concentrations of cell-free RNA. As demonstrated for two healthy donors in the electropherogram plots, cell-free RNA concentration is maintained to roughly draw time levels, even after 7 days ambient condition storage for the RNA Complete blood samples (Figure 6).
RNA Complete BCT formulation limits increases in cfRNA during ambient temperature storage.
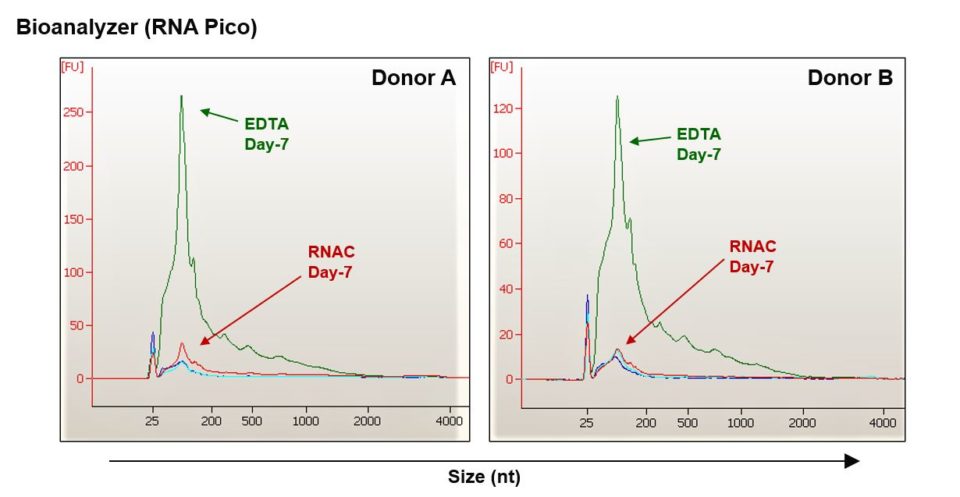
Figure 6
How can this knowledge be translated into usable data? For this, we developed an in-house plasma RNA-seq protocol. Briefly, blood was collected and stored for up to 7 days. From here, plasma was collected and cell-free RNA isolated, per our previous workflow. Total cell-free RNA, equating to roughly 10 nanograms, is then used in sequencing library preparation using a commercially available kit, which is dependent on an upfront ribosomal RNA depletion step. From here, each library is quantitated via qPCR methods and sized via TapeStation analysis. Finally, libraries are pooled and sequenced on an Illumina NextSeq. All data is saved and analyzed using the cloud-based Illumina BaseSpace server.
We first looked at total library yield, as this is dependent on cell-free RNA input. We expected library yield to increase in the EDTA samples, but remain constant in the RNA Complete samples. This is exactly what we observed. RNA Complete samples maintained draw time cell-free RNA levels, and thus maintained overall library yield.
“RNA Complete samples maintained draw time cell-free RNA levels, and thus maintained overall library yield.”
Library size should maintain the 300 to 400 nucleotide size specified by the manufacturer. And this was observed for both the EDTA and RNA Complete samples. With TapeStation analysis, we again observed a dramatic increase in library yield for the EDTA samples, whereas the RNA Complete samples maintained a library yield to draw time levels. From here, each library sample was sequenced and then aligned to the referenced human genome. With EDTA samples, we observed a dramatic reduction in sequencing reads aligning to critical coding sequences, such as messenger RNA. Importantly, coding sequence reads were retained in samples drawn into the RNA Complete BCT (Figure 7).
Samples stored in RNA Complete retain sequence alignment distribution to draw time levels.
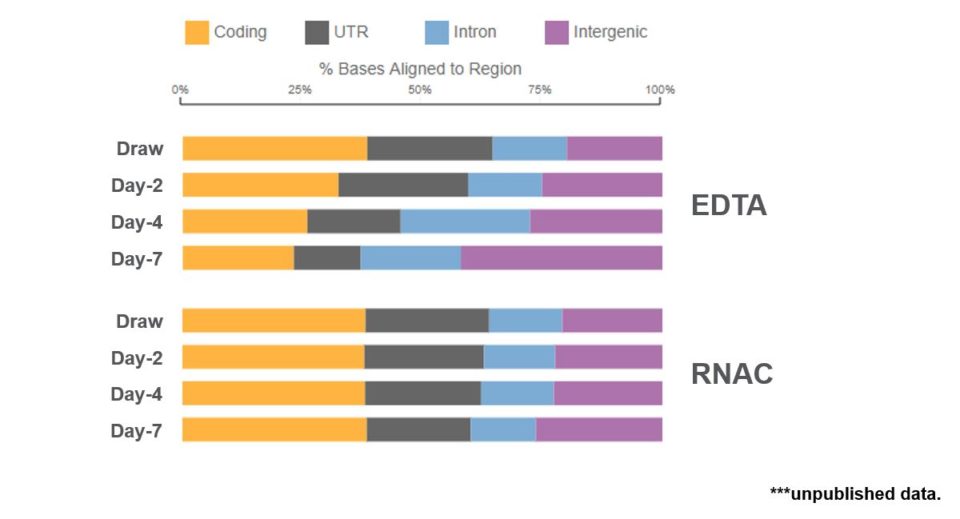
Figure 7
Differential gene expression analysis was then used to determine differences between EDTA and RNA Complete samples at draw time. In a five donor set, no significant differences in gene expression were noted between EDTA and RNA Complete BCTs. This is useful and demonstrates draw time performance is equivalent between the two types. Total assessed gene count between the two was roughly 12,450 genes. Many of the most abundant transcripts observed in draw time samples are red blood cell specific transcripts, as denoted by the red arrows in the right panel (see video for reference).
From here, we used general differential expression analysis to determine how well RNA Complete functioned to maintain draw time plasma transcriptome. For EDTA samples in a five donor set, roughly 950 genes changed by more than two fold over the course of 7 days ambient temperature storage. A majority of these demonstrated increased abundance with many, such as the globin transcripts, coming from red blood cell sources. Compared to EDTA samples, the same donors drawn into RNA Complete maintain a much tighter expression level out to 7 days. Here, only eight genes demonstrated greater than two fold change. And importantly, all eight of these genes were low expressing transcripts (Figure 8).
Draw time plasma transcriptome is maintained in RNA Complete samples for up to 7 days when samples are stored at ambient temperature.
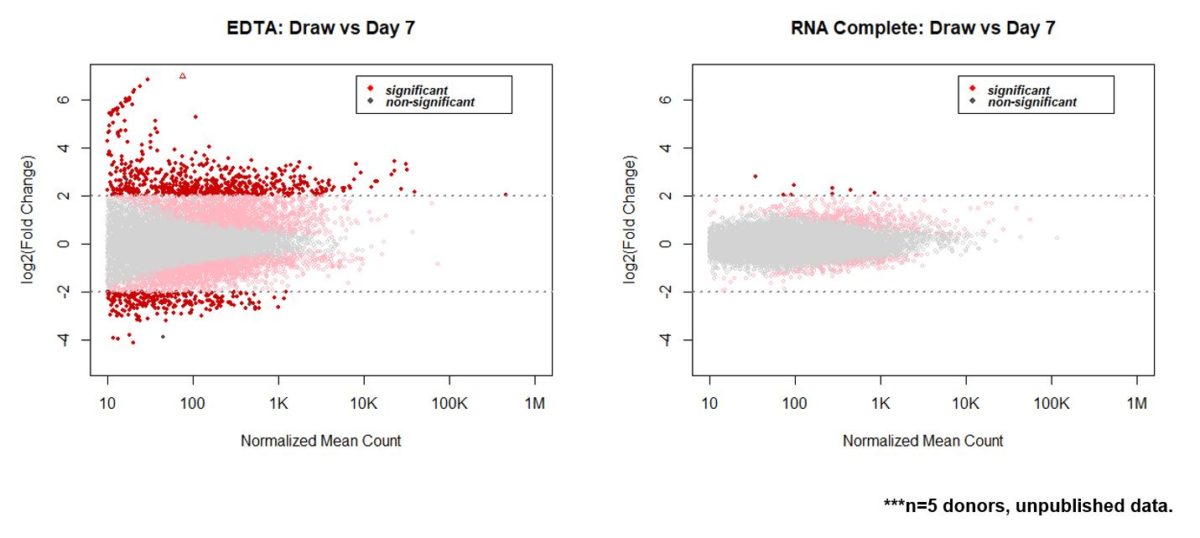
Figure 8
In conclusion, RNA Complete maintains the draw time plasma transcriptome for up to 7 days when samples are stored at ambient temperature. This is critical for next generation sequencing assay developers, because dilution of target transcripts by non-specific increases in library yield, for example, bi-cellular breakdown and disease non-specific cell-free RNA increases, can and will decrease the sensitivity of the particular NGS assay.
In conclusion, we’ve developed a blood collection tube that, at ambient storage temperature, limits hemolysis and prevents changes in plasma volume. It limits increases in extracellular vesicles and maintains draw time cell-free RNA concentration, thus limiting changes in the plasma transcriptome, as demonstrated with our RNA-seq workflow. The Streck RNA Complete BCT is useful to those interested in assay of extracellular vesicles and cell-free RNA, and is directly applicable to sample stabilization for plasma RNA-seq workflows.
Where are we going with our further directions with RNA Complete BCT? We are currently investigating stabilization of draw time levels of microRNAs. We’ve demonstrated this with messenger RNA in our RNA-seq workflows, and we’re currently testing microRNA panels with this blood collection tube. Finally, we are profiling plasma extracellular vesicles using surface marker expression to gauge how well the RNA Complete BCT maintains different subpopulations of extracellular vesicles, such as microvesicles and exosomes.
I thank you for your attention. For further questions, please contact Streck or visit the RNA Complete BCT webpage at the following web address. Thank you.

Dr. George received his Ph.D. in Cancer Biology from the University of Nebraska Medical Center and joined Streck in 2015. His current research and development interests include facilitating research into liquid biopsy and the multitude of pre-analytical variables associated with sample collection, processing and analysis.
Cell-Free DNA BCT and RNA Complete BCT are for Research Use Only. Not for use in diagnostic procedures in the U.S. A CE version of these tubes is also available. Cell-Free DNA BCT CE and RNA Complete BCT CE are for Export Only. Not for sale in the U.S.


FDA clearance brings liquid biopsy into a new era