Antimicrobial Resistance: “The Silent Pandemic”
Topics Featured
Bloodstream infections (BSIs) are the leading cause of mortality from infection. This is primarily due to growing antimicrobial resistance (AMR), which occurs when pathogens develop mechanisms to avoid being killed by antimicrobial treatments. According to the 2019 Antibiotic Resistance (AR) Threats Report from the Centers for Disease Control and Prevention, more than 2.8 million antibiotic-resistant infections occur in the United States each year, and more than 35,000 people die as a result. When considering which pathogens develop AMR, bacteria are the most common, with several species displaying resistance to multiple antibiotics. However, reports of resistant fungi have increased in recent years.
How does a hospital diagnose BSI?
For several years, blood culture has been the primary means of identifying organisms responsible for disease. Blood culture involves the growing of bacteria or fungi present in the blood with subsequent identification based on phenotypic antimicrobial susceptibility. However, this method of detection is very time consuming, and when it comes to BSI, a few hours can mean life or death. New diagnostic assays have been developed which provide more rapid detection of bacteria and yeast in the blood — among these is PCR-based analysis, which directly detects pathogens in positive blood cultures.
Though PCR-based assays have benefits in terms of time and effort, limitations exist. Blood is complex in composition and may include trace nucleic acids that could confound accurate results. Further, while the overall process is quicker than traditional culture-based methods, preparation of blood samples for PCR analysis involves several steps which are subject to experimental error. Because proper identification of the disease-causing organism is integral to patient treatment, optimizing these steps is critical.
Molecular controls are a powerful tool to verify analytical technique. A proper control allows for optimization of all steps of sample processing and ensures that the readout is correct. In the case of PCR-based analysis, this control should meet the following criteria:
- It should easily integrate into the normal protocol.
- It should have similar composition to usual samples.
- It should contain the nucleic acids being targeted by PCR.
By using a control that meets these standards, lab personnel can be confident in their sample preparation and analysis.
MDx-Chex® for BCID2, MDx-Chex® for BC-GP and MDx-Chex® for BC-GN are patient-like full process controls designed to validate the entire analytical process of the BCID2, BC-GP or BC-GN Panels on the BIOFIRE® (BCID2) or Luminex VERIGENE® (BC-GP, BC-GN) systems, including:
- Cell lysis
- DNA extraction
- Detection
- Analysis
How do pathogens resist antibiotics?
To understand how antibiotic resistance occurs, we first need to learn how antibiotics work.
While all antibiotics aim to kill bacteria or stop their growth, they can be stratified based on the specific bacterial processes and parts targeted. Antibiotic classes like penicillins and cephalosporins target the bacterial cell wall or membrane, preventing cell division or disrupting the structure of the bacteria. Other antibiotics target cellular processes, such as protein synthesis (macrolides, tetracyclines) or DNA replication (fluoroquinolones), restricting bacterial survival.
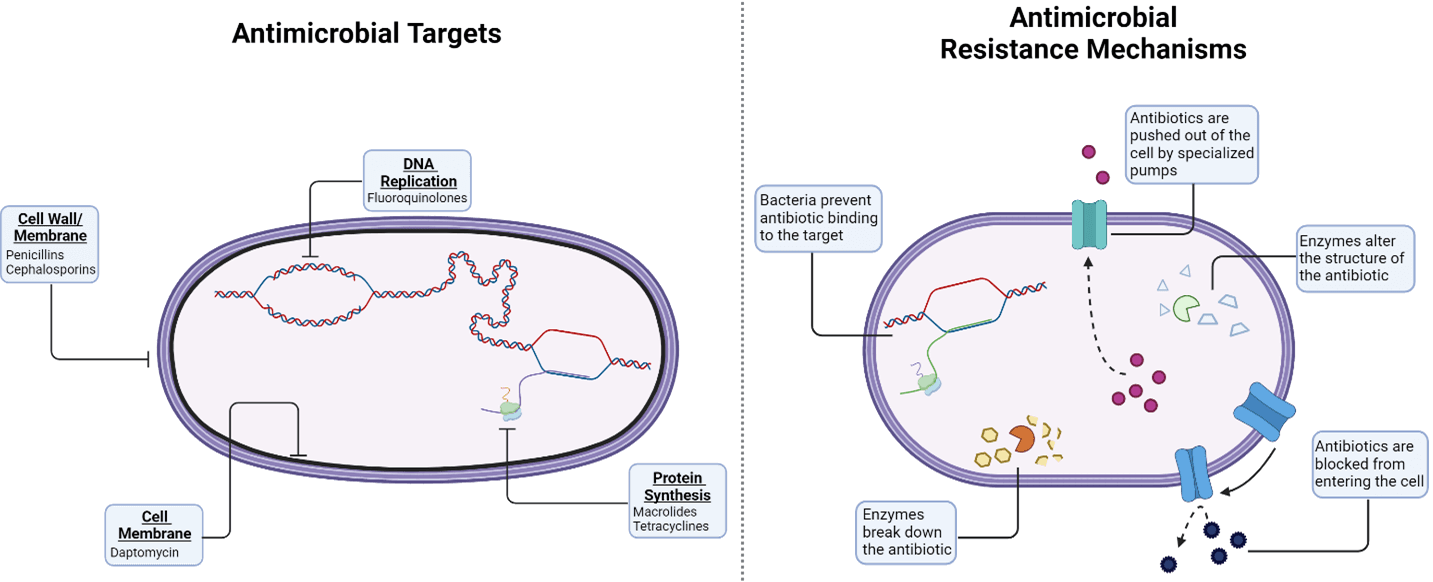
But bacteria are expert evolvers, making them a constantly moving target for antibiotics. Over time, bacteria have developed clever ways to avoid being killed. To defend against attacks to the cell wall or membrane, certain bacteria can restrict access the cell by preventing internalization of the antibiotic. Others produce factors that modify the antibiotic to prevent its binding to the bacterial surface. Antibiotics aren’t safe once they reach the inside of the cell, either. Several bacteria have mechanisms to prevent antibacterial action, including changing the structure of the antibiotic target to prevent binding, releasing enzymes that destroy the antibiotic and actively pumping the antibiotic out of the cell.
The ability of bacteria to evolve becomes a greater issue when we consider that AMR mechanisms can be transferred from bacteria-to-bacteria through several different types of gene transfer. This means that as few as one antibiotic-resistant bacterium can cause widespread resistance in the population.
Where does AMR spread?
In addition to transfer between bacteria, AMR can become a major issue if it is spread throughout hospitals, the community or the environment. AMR often develops in healthcare facilities, as this is primarily where antibiotics are used for treatment. Once antibiotic-resistant bacterial populations arise, they can spread to unsuspecting patients and clinicians through vectors such as medical devices, inappropriate infection control actions and hospital plumbing.
In the community, antibiotic resistant bacteria can spread from person to person during mundane activities, such as shaking hands or working out. Even spending time with your pet makes you vulnerable to possible infection with antibiotic-resistant bacteria if you aren’t properly washing your hands after interacting with them. AMR can become a global issue when people travel internationally, as they can become infected with antibiotic-resistant bacteria from other people, animals, contaminated food or water and spread the infection upon return.
What bacteria are a threat?
In the 2019 AR Threats Report, the CDC defined the top 18 pathogens that require our attention, ranking the threat of each as ‘urgent,’ ‘serious’ or ‘concerning.’
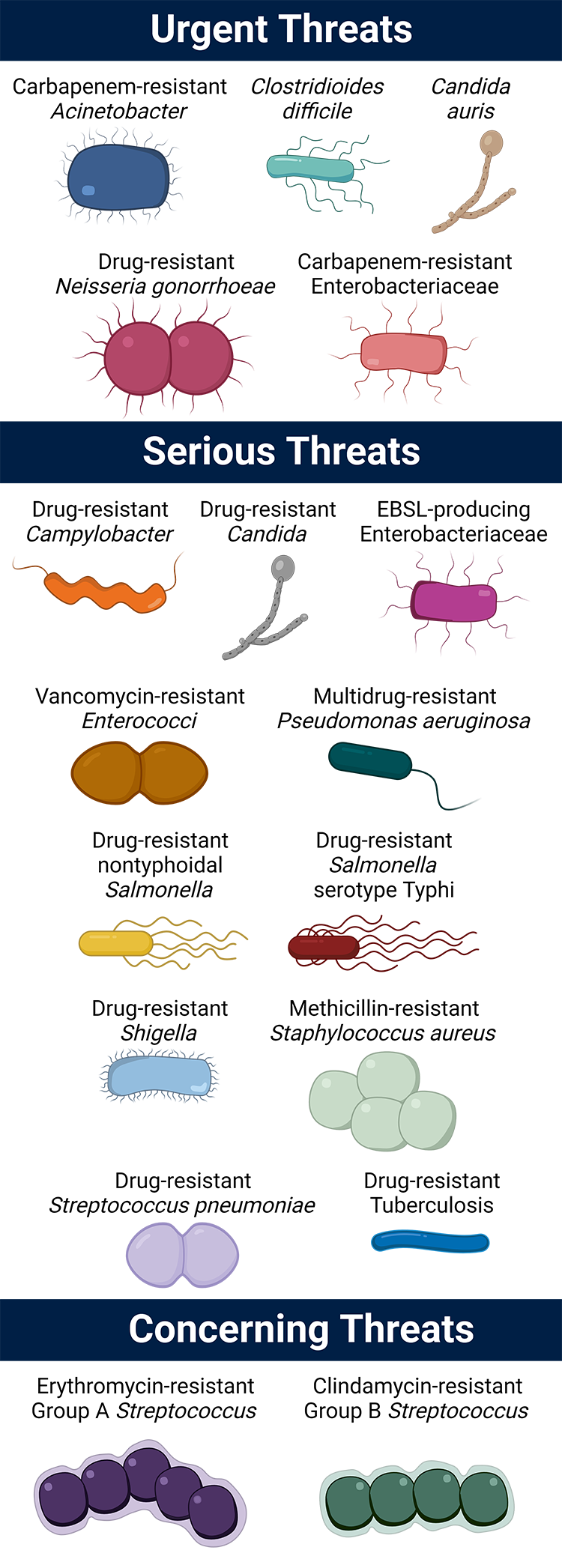
Urgent threats
- Carbapenem-resistant Acinetobacter
- Candida auris (C. auris)
- Clostridioides difficile (C. difficile)*
- Carbapenem-resistant Enterobacteriaceae (CRE)
- Drug-resistant Neisseria gonorrhoeae (N. gonorrhoeae)
Serious threats
- Drug-resistant Campylobacter
- Drug-resistant Candida
- Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae
- Vancomycin-resistant Enterococci (VRE)
- Multidrug-resistant Pseudomonas aeruginosa (P. aeruginosa)
- Drug-resistant nontyphoidal Salmonella
- Drug-resistant Salmonella serotype Typhi
- Drug-resistant Shigella
- Methicillin-resistant Staphylococcus aureus (MRSA)
- Drug-resistant Streptococcus pneumoniae (S. pneumoniae)
- Drug-resistant Tuberculosis (TB)
Concerning threats
- Erythromycin-resistant Group A Streptococcus (GAS)
- Clindamycin-resistant Group B Streptococcus (GBS)
*C. difficile is not itself antibiotic-resistant. Rather, C. difficile infection is associated with antibiotic use, and thus, commonly arises when treating these threats.
What can we do?
Antibiotic-resistant infections in humans can be difficult, and sometimes impossible, to treat. People receiving healthcare or those with weakened immune systems are often at higher risk for getting an infection.
In 2020, AMR monitoring decreased, as most healthcare professionals focused on responding to the COVID-19 pandemic. However, AMR didn’t slow down — in fact, the threat of antibiotic-resistant infections has gotten worse. As COVID-19 cases steadily decrease, the CDC has returned its attention to AMR.
CDC efforts focus on five main ways to combat antimicrobial resistance:
- Preventing Infections
- By preventing infections before they start – through washing your hands, getting vaccinated, using antibiotics appropriately – we can keep AMR at bay.
- Antibiotic/Antifungal Use
- Antibiotic stewardship promotes the use of antibiotics and antifungals only when necessary and per recommendations for prescribing practice.
- Vaccines, Therapeutics and Diagnostics
- Treatment following infection is not the only way to stop the spread of antibiotic-resistant bacteria. Prevention products, like vaccines, can help stop infections before they happen.
- Environment and Sanitation
- With increased efforts to monitor wastewater, including the establishment of the National Wastewater Surveillance System, we can better understand how AMR spreads through the environment.
- Tracking and Data
- If we know how and where AMR is occurring, then we can better devise ways to prevent infection and slow the spread of resistance.
One way hospital infection control staff can check to see which resistance mechanisms are present in their community or healthcare system is to use an antibiotic resistance surveillance method, such as Streck ARM-D® Kits. These kits allow lab personnel to detect the most clinically important β-lactamases for current and emerging threats, including carbapenem-resistant Enterobacteriaceae (CRE), ampCs, extended-spectrum beta-lactamases (ESBL) and mobilized colistin resistance (mcr).

Analyze with ease and certainty.
With full-process controls and antibiotic resistance kits, Streck has the products to help you streamline your molecular diagnostics testing, aid antibiotic stewardship and ensure positive patient outcomes.
Streck ARM-D Kits are for Research Use Only. Not for use in diagnostic procedures.
New antibiotic fights antimicrobial resistant bacteria