ePoster 759 | AACR 2020
Streck Research and Development, La Vista, NE
RNA Complete BCT™: A novel blood collection tube targeting circulating RNA and extracellular vesicles
Nicholas George, Ph.D., Lisa Bartron, and Eunju Seong, Ph.D.
For Research Use Only. Not for use in diagnostic procedures in the U.S.
Abstract
Precision medicine, specifically in medical oncology, has been significantly enabled and the pace accelerated with the advent of liquid biopsy-based diagnostics. While liquid biopsy assays targeting circulating cell-free DNA (cfDNA) provide useful diagnostic information such as mutational burden, they are limited to a static assessment of the disease state due to cfDNA derivation from dying cells. Assays exploiting circulating extracellular vesicles (EVs) and EV-associated cargo, such as cell-free RNA (cfRNA), provide a comprehensive and dynamic view of the disease as these analytes derive from both living and dying cells. A significant hurdle in making liquid biopsy testing with EVs and cfRNA commonplace is donor specimen integrity. Collected patient blood samples undergo time-dependent degradation leading to increases in disease non-specific EVs and cfRNA thereby compromising the accuracy and reproducibility of analyte measurement. To overcome this vulnerability we have developed the Streck RNA Complete BCT (RNAC), a first of its kind blood collection tube intended for stabilizing draw time concentrations of extracellular vesicles and cell-free RNA. Sample integrity will benefit users interested in direct analysis of EV populations and indirect analysis of cfRNA through RT-PCR or next-generation sequencing-based methods. Incorporation of this blood collection tube into the pre-analytical workflow will allow long distance shipping, batch and/or delayed sample processing without effect on sample integrity or downstream analyte analysis.
Materials & Methods
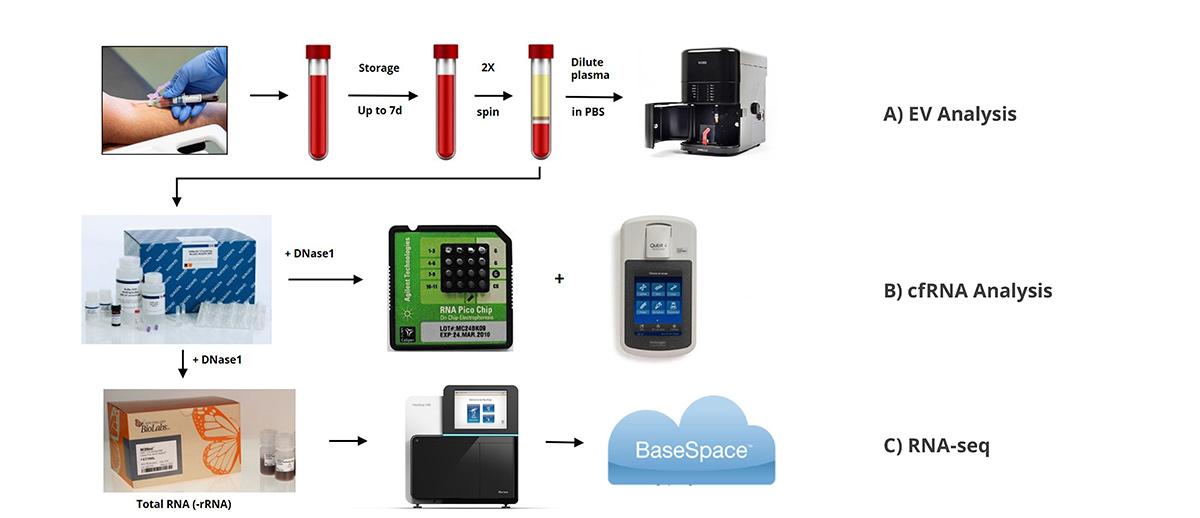
Plasma Sample Preparation: Blood samples were collected from healthy donors via venipuncture in accordance with IRB #830575 (Creighton University, Omaha, NE). Plasma was isolated using a general double spin protocol (1800 g x 15 min & 2800 g x 15 min) and frozen at -80 oC until use.
EV Analysis: Assay of EV concentration and particle size utilized the NanoSight NS300 instrument (Malvern Panalytical Ltd.). Frozen plasma samples were rapidly thawed, diluted 1:50-1:500 in PBS, and assayed per manufacturer’s recommendation.
cfRNA Isolation and Analysis: Plasma samples were rapidly thawed and total nucleic acid was isolated using the QIAamp Circulating Nucleic Acid Kit according to manufacturer’s recommendations miRNA protocol (Qiagen Inc.). Sample lysis was increased from 30 min to 60 min. Carryover DNA was depleted and cfRNA concentrated using the RNase-free DNase Set and RNeasy MinElute Cleanup Kit (both from Qiagen). Cell-free RNA analysis utilized the 2100 Bioanalyzer with the RNA 6000 Pico kit (Agilent Technologies, Inc., Santa Clara, USA) and Qubit RNA HS Assay Kit (Thermo-Fisher), both according to manufacturer’s recommendation.
RNA-seq: Cell-free RNA was depleted of rRNA using the NEBNext rRNA Depletion Kit and then used in sequencing library preparation with the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina, both according to provided instructions. Libraries were quantified (NEBNext Library Quant Kit for Illumina) and size determined using the High Sensitivity D1000 ScreenTape run on the 4200 TapeStation system (Agilent). Libraries were pooled and sequenced on the Illumina NextSeq 550 using High Output v2.5 sequencing kit (Illumina, San Diego, CA). All data were output and saved to the cloud-based BaseSpace server (Illumina). Reads were aligned to the human genome and differential expression was completed using the BaseSpace applications Top-Hat Alignment and DESeq2, respectively.
Results
Maintenance of Draw Time Concentrations of Extracellular Vesicles
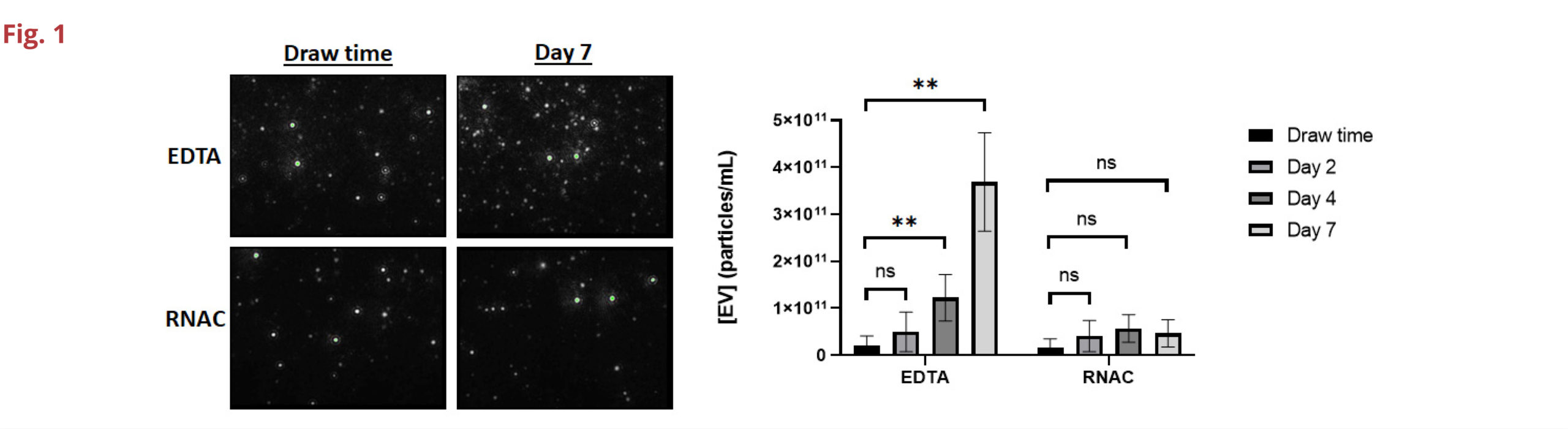
Fig 1. Concentrations of extracellular vesicles are maintained in RNAC samples for up to 7 days when stored at ambient temperature. Nine different donors were drawn into both EDTA and RNAC BCTs and plasma was isolated immediately (draw time) or after 2, 4, and 7 days ambient condition storage. The mean overall particle size remained unchanged during this time (data not shown). (Paired sample t-test, ns = not significant, ** = p≤0.01).
cfRNA Concentration is Maintained in RNAC for up to 7 Days
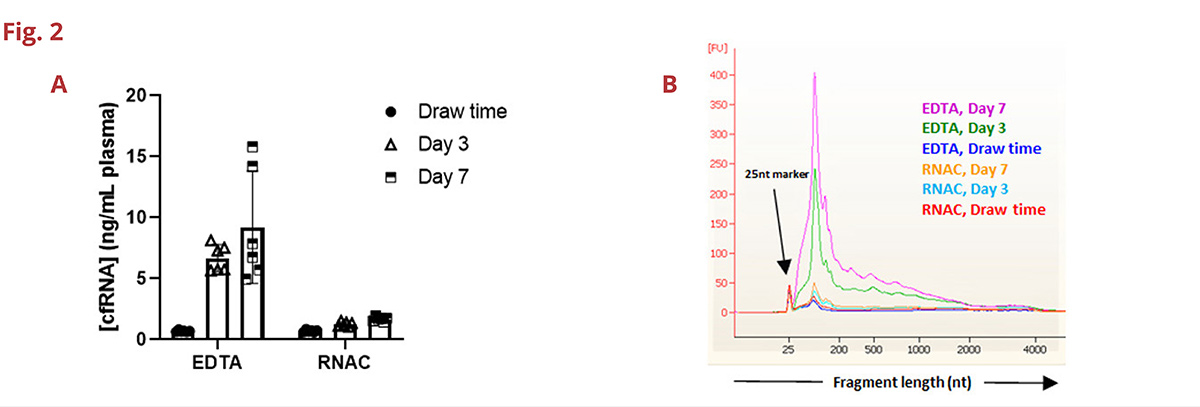
Fig 2. A. Cell-free RNA concentration is maintained in RNAC out to 7 days when stored at ambient temperature (Qubit assay, n=6 healthy donors). B. Bioanalyzer traces of cfRNA isolated from a single donor demonstrate a time-dependent increase in small RNA species in blood drawn into EDTA BCTs. Day 3 and Day 7 cfRNA concentration is maintained to near draw time levels for the same donor drawn into RNAC.
Sequencing Library Yield is Conserved in RNAC Out to 7 Days (Ambient Storage)
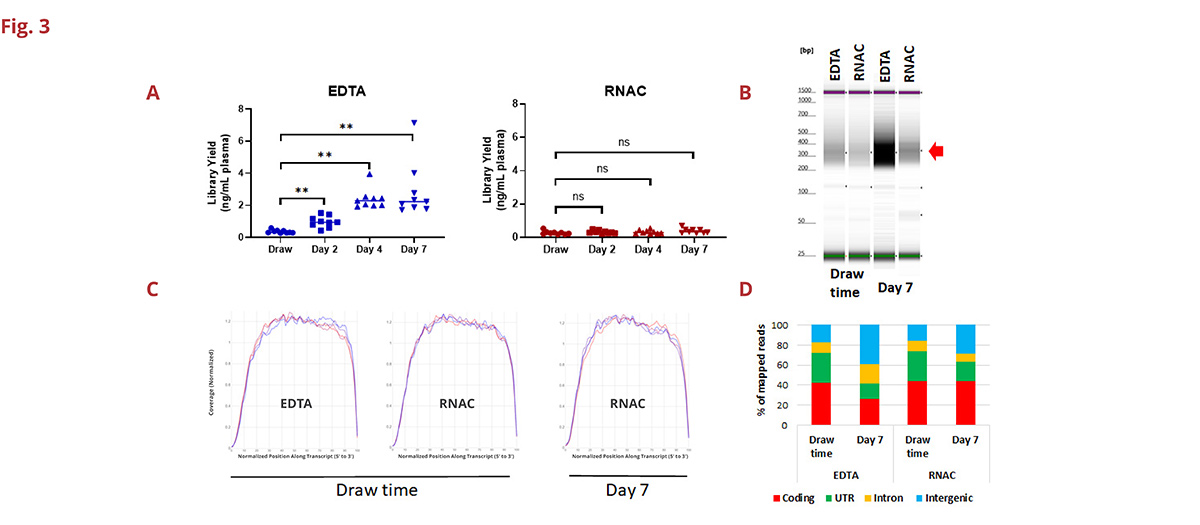
Fig 3. A. Overall sequencing library yield is directly related to the input amount of cell-free RNA. While cell-free RNA levels increase as a function of ambient condition storage time in EDTA samples, so does overall library yield. The same donors drawn into RNAC BCTs maintain library yield to draw time levels (paired sample t-test, ns = not significant, ** = p < 0.01, n=9 donors). B. Relative sequencing library size and yield for a representative healthy donor (composite gel image demonstrating the expected ~350nt fragment size for each library (red arrow), this increases in EDTA but maintains baseline level in RNAC). C. Transcript coverage for individual libraries is similar between EDTA and RNAC samples and does not change over the course of 7-day ambient condition storage. D. Mapped reads aligning to coding sequence decrease with time in blood samples drawn into EDTA BCTs and remain constant in the same donors drawn into RNAC (averaged reads from a nine donor set).
Maintenance of Draw Time Plasma Transcriptome in RNAC
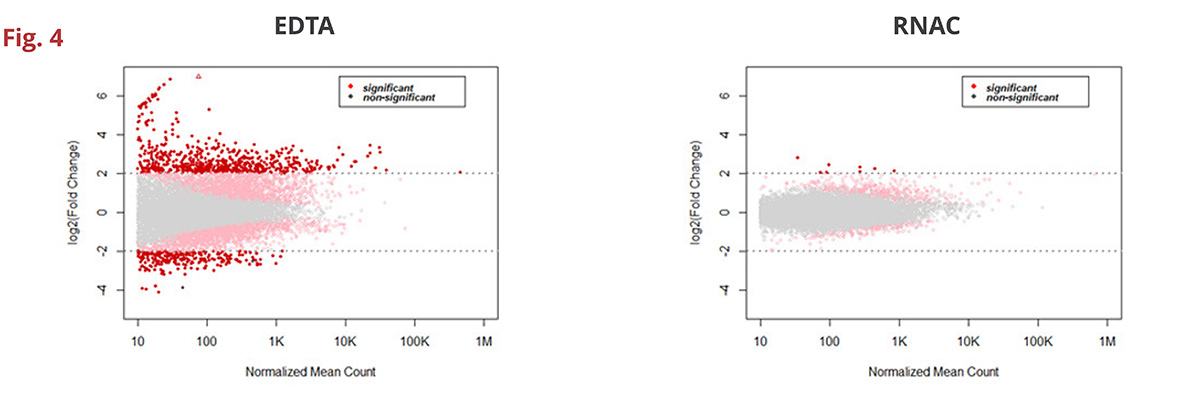
Fig 4. Changes in gene expression of greater than log2-fold are minimized in RNAC – only 8 genes demonstrate changes compared to 928 in EDTA BCTs (n=5 donors). Importantly, highly abundant transcripts (i.e., those with a higher normalized mean count) remain at near baseline levels throughout 7 days of ambient condition storage.
Conclusions
- Streck RNA Complete BCT maintains draw time concentrations of plasma extracellular vesicles for up to 7 days when blood is stored under ambient conditions.
- Plasma cell-free RNA isolated from RNA Complete BCT can be used in generic RNA-seq workflows and major parameters (transcript coverage, mapped reads, draw time plasma transcriptome) do not vary from gold-standard EDTA BCTs immediately after blood draw.
- RNA Complete BCT maintains draw time levels of cfRNA and thus resultant sequencing library yield for up to 7 days when blood is stored at ambient conditions.
- Plasma transcriptome is maintained in Streck RNA Complete BCT out to 7-day ambient condition storage.
