How Nanotrap® Technology for Wastewater Surveillance Improves SARS-CoV-2 Workflows
Associate Professor and Graduate Program Director for the Department of Civil and Environmental Engineering, Caitlyn Butler, shares how Nanotrap particle technology improved her SARS-CoV-2 wastewater surveillance workflow at the University of Massachusetts, Amherst.
In this interview, Caitlyn Butler, from the University of Massachusetts, Amherst, discusses how Nanotrap particle technology enables her lab to obtain consistent and accurate results from samples collected across 43 sites on the campus. With a higher RNA yield and a shorter processing time, University of Massachusetts, Amherst has been able to ramp up the amount of samples they test daily.

Tell us about your current research at University of Massachusetts Amherst and your ultimate goals.
CB: For the last two years our lab has been engaged in service-based research as we work with University of Massachusetts Amherst on monitoring SARS-CoV-2 in wastewater on campus as part of a comprehensive COVID-19 response plan. We are currently collecting wastewater samples around campus from 43 sample sites three times a week. Our mission in collecting these data is to inform the campus health response to SARS-CoV-2 local trends. We do this by providing the University with the information and data we are collecting so they can ensure that safe campus health decisions are being made to protect the students and faculty. While a goal of ours is to support a safe and healthy campus, we are also taking the opportunity to ask emerging research questions associated with the expansion of wastewater-based epidemiology. Our guiding question is, what do we need to do to improve the overall efficacy of wastewater-based epidemiology for extending communities and organizations?
Please share with us more about your workflow and your metrics for success.
CB: A broad overview of our workflow starts with the acquisition of samples across our 43 sites on campus. We use Ceres Nanosciences’ Nanotrap Magnetic Virus Particles to isolate SARS-CoV-2 , which we do through a series of steps as we use a passive sampling approach. After we use the Nanotrap technology for capture and concentration, we send the extracted RNA to our campus clinical testing center for qPCR. We adapted our RNA concentration and isolation protocol to interface with a clinical qPCR protocol and pipeline to allow us to collaborate with our on-campus clinical test center and to expand our sample processing capacity. Nanotrap particles were integral to the concentration and isolation procedure that allowed this collaboration and integration with a clinical testing assay. We define our metrics of success on timeliness of sample preparation, timeliness of data acquisition, accuracy of the results obtained, and the consistency of the data we collect.
“With the Nanotrap technology, I can now increase the capacity of samples I am able to process in my workflow and provide enriched information on SARS-CoV-2 presence to the University of Massachusetts Amherst campus.”
Please describe how the solution in your lab is aiding you in doing more today.
CB: Using Nanotrap technology in the lab has been imperative for increasing the number of samples being processed in our pipeline. Prior to using this technology, we were using filtration, which was proving to be a bottleneck in our process that we needed to overcome, as it limited the number of samples we could process daily. By adopting Nanotrap technology, this allowed us to quickly scale up the volume of samples for processing. The technology has also greatly propelled us forward in our own protocols.
What impact does using Nanotrap technology have on public health response/campus health response?
CB: Using the Nanotrap technology, we developed a modified protocol that is helping us serve more sites on campus. This workflow has helped us improve and enrich the information we were already receiving, because we were able to expand the amount of sample sites we were monitoring. We are processing a greater number of samples in the same amount of time since we have been using Nanotrap particles, because they removed the time barrier that was built by our use of filtration in our original wastewater-based epidemiology efforts. Serving more sites allows us to build a more comprehensive COVID-19 profile of the campus.
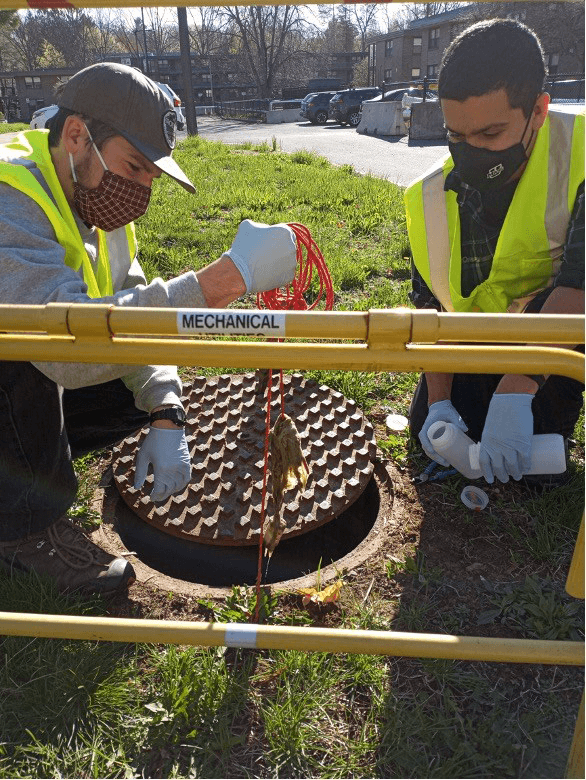
Two members of the University of Massachusetts, Amherst wastewater surveillance team collecting a sample for the lab from one of the on-campus collection sites.

Member of the University of Massachusetts, Amherst SARS-CoV-2wastewater surveillance team working with samples in the lab.
With SARS-CoV-2 wastewater surveillance being a focus in many labs, how are you addressing it today in your lab, and what are your future goals with overall wastewater monitoring?
CB: While the COVID-19 pandemic transitions into epidemic status, we are using this time to pivot from solely SARSCoV-2 wastewater surveillance and diversify what we are doing with wastewater-based epidemiology in the lab. There is a lot of opportunity for us to learn what the information collected from wastewater surveillance methods can do when it comes to improving the quality of public health and general community health decisions. As we continue to test wastewater for SARS-CoV-2, we also plan to soon expand our workflow and explore a suite of viruses and other biomarkers in the future.
