#P-117 | APHL 2019
Streck Research and Development, La Vista, NE
Detection of Mobilized Colistin Resistance (mcr) Genes by Multiplex Real-Time PCR: Improving Surveillance of an Emerging Threat
E. Alao; M.P. Torres; S. Cossette; C. Connelly
Abstract
Background: Recent increases in colistin-resistant infections led the CDC to launch an urgent public health response for mcr surveillance. Colistin is a last resort antibiotic that is more utilized due to increases in carbapenem-resistant infections. Since mcr-1 was first reported, more mcr gene variants have been identified, but few screening tools have been developed to rapidly detect mcr-positive samples. To improve surveillance for mcr genes, we describe a multiplex real-time PCR assay that detects mcr gene families 1 through 5 in less than 45 minutes.
Materials/Methods: This study utilizes sequence-specific primers and probes for the real-time PCR-based detection of mobilized colistin resistance mcr variants. An internal control (IC), targeting a conserved region in Gram-negative bacteria, is also included in the multiplex mix to discriminate false negative samples. Positive DNA controls are included with the multiplex assay. Data was generated using the Bio-Rad® CFX96 Touch™ Real-time PCR Detection System.
Results: The mcr real-time PCR multiplex assay is optimized to amplify mcr families 1 through 5. Amplification of serial dilutions of target controls generated PCR efficiencies ≥ 99% and correlation coefficients ≥ 0.998. The sensitivity and specificity of the assay was evaluated using 90 clinical isolates and determined to be 100%. Internal control DNA were detected in 100% of samples and within 20 PCR cycles.
Conclusions: Streck ARM-D® Kit, MCR provides a rapid amplification and detection strategy to monitor plasmid-mediated colistin resistance genes. The data demonstrate a sensitive and specific assay, with no observed cross-reactivity with previously characterized clinical isolates from Gram-negative organisms. The results demonstrate this assay can serve as a screening tool for surveillance of mcr-mediated colistin resistance, thereby improving antimicrobial stewardship practices to minimize mcr gene dissemination into the community.
Background
- Global increase in carbapenemase-producing Enterobacteriaceae has resulted in increased use of colistin.
- Colistin is considered a last resort antibiotic for multi-drug resistant Gram-negative bacteria.
- Mobilized colistin resistance gene mcr-1 was first reported in 2015 in Escherichia coli strain isolated from pigs and humans in China.
- During the last 4 years mcr genes have been detected in over 30 countries and other types of bacteria such as Klebsiella pneumoniae and Salmonella enterica.
- According to the CDC, in the U.S. alone, over 50 human isolates have been reported across 19 states.
Figure 1: Tracking mcr in the U.S.
The following map shows where the mcr gene has been reported in U.S. human and food animal sources as of Nov. 2, 2018.
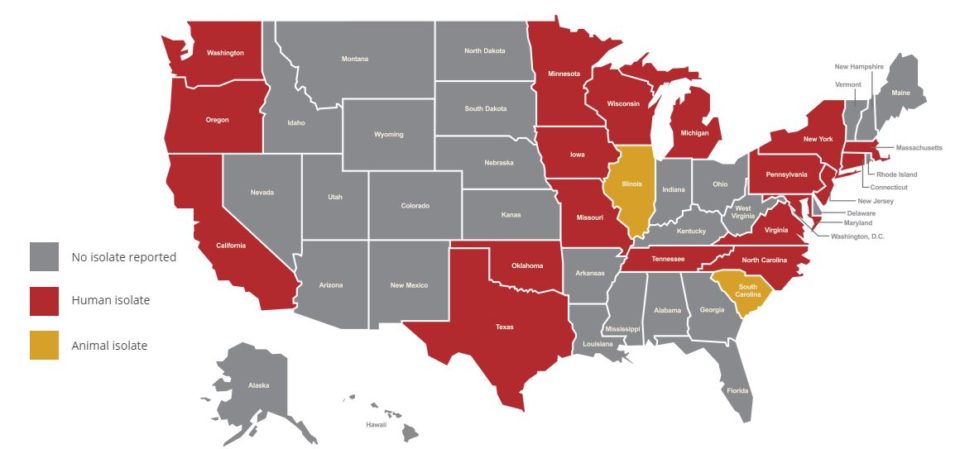
Map sourced from this CDC website
Results
The results in Figure 2 show the amplification of serial dilutions of ARM-D Kit, MCR Control Mix targets. Standard curves are shown for each target control. PCR efficiencies were over 99% for all the target controls and correlation coefficients were over 0.998. The amplification of the Multiplex Control Mix is shown in Figure 3 and respective Cq values for target controls are shown in Table 1.
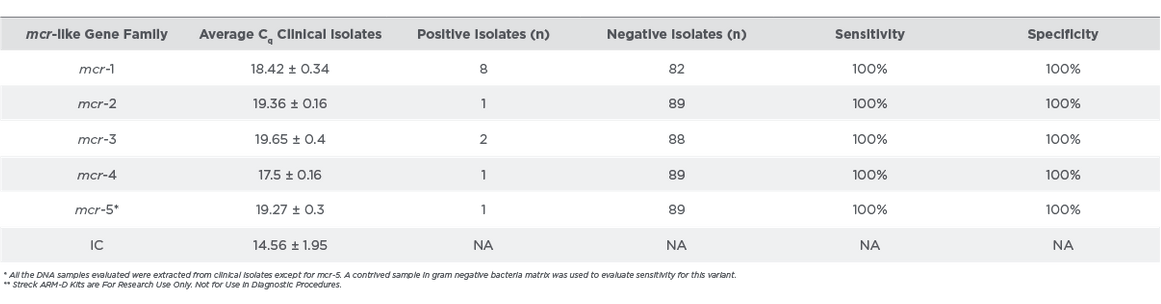
Out of the 90 isolates evaluated, 12 expressed mcr gene families. Representative amplification data of mcr-positive isolates with respect to positive controls is shown in Figure 4. The mcr positive isolates amplified within 20 cycles of the PCR run (Table 2). The clinical isolates were correctly identified by the mcr assay and the sensitivity and specificity were 100%.
Figure 2:
Real-time PCR amplification of serially-diluted Multiplex Control Mix of Streck ARM-D Kit, MCR. Standard curves show corresponding efficiencies and correlation coefficients for each target control, respectively.
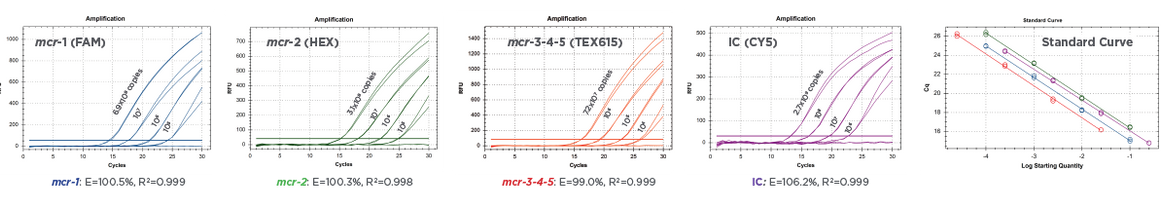
Figure 3:
Real-time PCR amplification of Streck ARM-D Kit, MCR Multiplex Control Mix
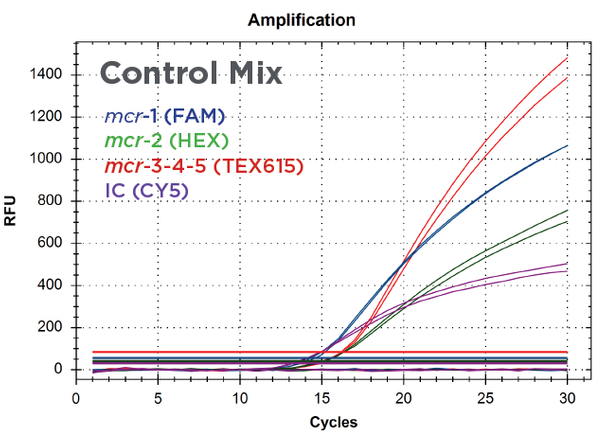
Table 1:
Cq Values Streck ARM-D Kit, MCR Control Mix
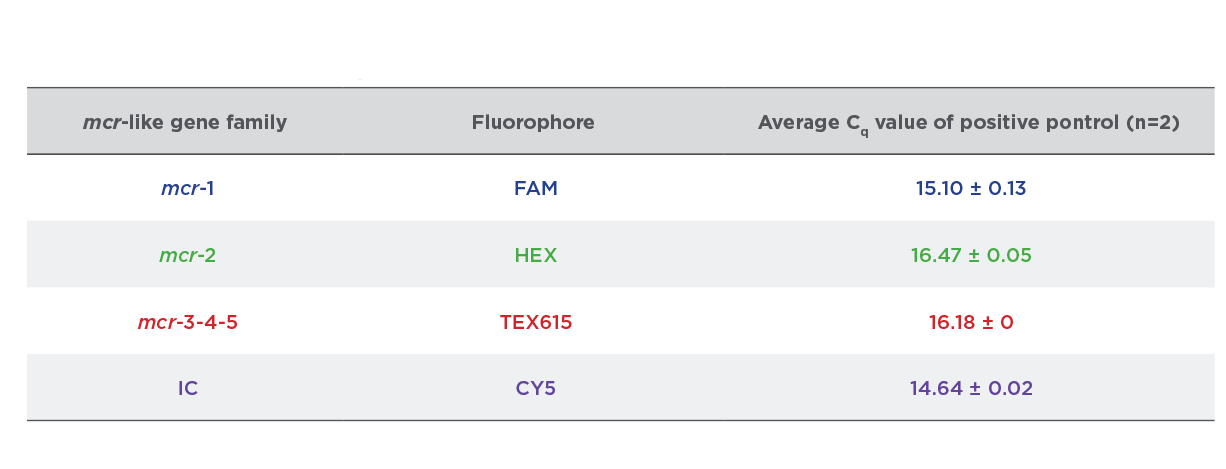
Figure 4:
Real-time PCR amplification of clinical isolates with Streck ARM-D Kit, MCR (n=90). Representative amplification plots of mcr-positive clinical isolates and positive control for each gene are shown. The internal control (IC) was amplified for all strains tested.

Table 2:
Streck ARM-D Kit, MCR-Clinical Isolate Testing**
Materials & Methods
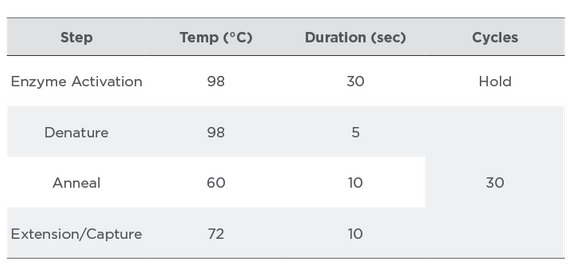
The genes covered in Streck ARM-D Kit, MCR were differentiated using target-specific hydrolysis probes, chemically linked to different fluorescent dyes. The gene variants covered by the assay are shown in Table 3. PCR master mix preparation and PCR cycling protocol are shown in Tables 4 and 5, respectively. The efficiency and correlation coefficients were determined for each target by the amplification of serial dilutions of the Control Mix. The sensitivity and specificity of the assay was evaluated with DNA purified from mcr-negative and mcr-positive overnight bacterial cultures (n=90).
Positive isolates for mcr gene targets were obtained from the CDC & FDA Antibiotic Resistance (AR) isolate bank panel with new or novel antibiotic resistance (AR Bank # 0346, 0349, 0350, 0493, 0494, 0495, 0496, 0497, 0538, 0539, 0540, 0635). DNA was isolated using the Qiagen® DNeasy® Blood and Tissue Kit or Exiprep Dx Bacteria Genomic DNA kit per the manufacturer’s instructions. Duplicate reactions were run for all clinical isolates and controls.
Table 3:
Gene targets covered by the Streck ARM-D Kit, MCR

Table 4:
Master Mix Preparation
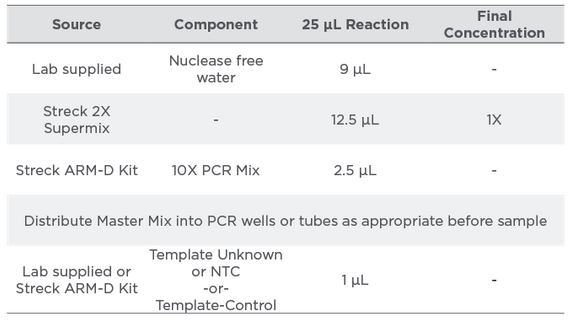
Table 5:
PCR Cycling Conditions
Conclusions
The Streck ARM-D Kit, MCR:
- Identifies and differentiates mcr-1, mcr-2 and mcr-3-4-5 gene families with 100% sensitivity and 100% specificity.
- Can detect up to 48 out of 51 mcr allelic variants within the described gene families.
- Represents a surveillance tool to track the spread of mobilized colistin resistance.
References
- Sun J, Zhang H, Liu YH, Feng Y, 2018. Towards Understanding mcr-like Colistin Resistance. Trends Microbiol. 26(9): 794-808.
- Caniaux I, van Belkum A, Zambardi G, Poirel L, Gros MF, 2017. mcr: Modern Colistin Resistance. Eur J Clin Microbiol Infect Dis. 36(3): 415-420.
- Karaiskos I, Souli M, Galani I, Giamarellou H, 2017. Colistin: Still a lifesaver for the 21st century? Expert Opin Drug Metab Toxicol. 13(1): 59-71.
- Centers for Disease Control and Prevention Tracking the mcr gene-Updated on Nov. 2, 2018. https://www.cdc.gov/drugresistance/biggest-threats/tracking/mcr.html
